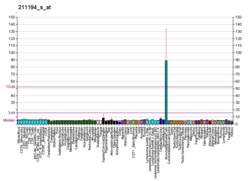
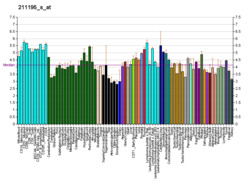
Tumor protein p63, typically referred to as p63, also known as transformation-related protein 63, is a protein that in humans is encoded by the TP63 (also known as the p63) gene.[5][6][7][8]
The TP63 gene was discovered 20 years after the discovery of the p53 tumor suppressor gene and along with p73 constitutes the p53 gene family based on their structural similarity.[9] Despite being discovered significantly later than p53, phylogenetic analysis of p53, p63 and p73, suggest that p63 was the original member of the family from which p53 and p73 evolved.[10]
Function
[edit]Tumor protein p63 is a member of the p53 family of transcription factors. p63 -/- mice have several developmental defects which include the lack of limbs and other tissues, such as teeth and mammary glands, which develop as a result of interactions between mesenchyme and epithelium. TP63 encodes for two main isoforms by alternative promoters (TAp63 and ΔNp63). ΔNp63 is involved in multiple functions during skin development and in adult stem/progenitor cell regulation.[11] In contrast, TAp63 has been mostly restricted to its apoptotic function and more recently as the guardian of oocyte integrity.[12] Recently, two new functions have been attributed to TAp63 in heart development[13] and premature aging.[14]
In mice, p63 is required for normal skin development via direct transcription of the membrane protein PERP. TP63 can also regulate PERP expression with TP53 in human cancer.[15]
Oocyte integrity
[edit]In oocytes, a unique quality control system has evolved that eliminates by apoptosis those oocytes in which chromosomes do not align correctly, or in which chromosomes cannot be repaired.[16] This monitoring system is conserved from fruit flies and nematodes to humans, and central to this system is the p53 protein family and, in vertebrates in particular, the p63 protein.[16] Oocytes that are unable to repair DNA double-strand breaks produced during meiosis by the process of homologous recombination are eliminated by apoptosis that is linked to p63.[16]
Clinical significance
[edit]At least 42 disease-causing mutations in this gene have been discovered.[17] TP63 mutations underlie several malformation syndromes that include cleft lip and/or palate as a hallmark feature.[18] Mutations in the TP63 gene are associated with ectrodactyly-ectodermal dysplasia-cleft syndrome in which a midline cleft lip is a common feature,[18] cleft lip/palate syndrome 3 (EEC3); ectrodactyly (also known as split-hand/foot malformation 4 (SHFM4)); ankyloblepharon-ectodermal dysplasia-cleft lip/palate (AEC) or Hay–Wells syndrome in which a midline cleft lip is also a common feature,[18] Acro–dermato–ungual–lacrimal–tooth syndrome (ADULT); limb-mammary syndrome; Rap-Hodgkin syndrome (RHS); and orofacial cleft 8.

Both cleft lip with or without a cleft palate and cleft palate only features have been seen to segregate within the same family with a TP63 mutation.[18] Recently, induced pluripotent stem cells have been produced from patients affected by EEC syndromes by cell reprogramming. The defective epithelial commitment could be partially rescued by a small therapeutic compound.[19]
Molecular mechanism
[edit]Transcription factor p63 is a key regulator of epidermal keratinocyte proliferation and differentiation. In a recent study, researchers used EEC-patient-derived skin keratinocytes carrying heterozygous p63 DNA-binding domain mutations as the cellular model to characterize the global gene regulatory alteration. The epidermal cell identity was compromised in p63 mutant keratinocytes. Besides, p63-binding loss and loss of active enhancers occurs at a genome-wide scale in patient keratinocytes carrying heterozygous EEC mutations. [20] Besides, using a multi-omics approach, the deregulated function of DNA loops mediated by p63 and CTCF represents an additional layer to the disease mechanism. It seems that a number of loci nearby epidermal genes were organized into a ‘regulatory chromatin hub’ within the chromatin interactions, mediated by CTCF in epidermal keratinocytes. Such hubs contain multiple connecting DNA loops that require not only CTCF binding that is rather static but also binding of cell type-specific TFs, like p63, for the transcriptional activity. In this model, p63 may be essential to make the DNA loops active in transcription. [21]
Vulvar cancer
[edit]TP63 has been observed overexpressed in Vulvar Squamous Cell Carcinoma samples, in association with hypermethylation-Induced inactivation of the IRF6 tumor suppressor gene. [22] Indeed, mRNA levels of TP63 tested higher in Vulvar cancer samples when compared with those of normal skin and preneoplastic vulvar lesions, thus underscoring an epigenetic cross-link between IRF6 gene and the oncogene TP63. [22]
Diagnostic utility
[edit]
p63 immunostaining has utility for head and neck squamous cell carcinomas, differentiating prostatic adenocarcinoma (the most common type of prostate cancer) and benign prostatic tissue;[23] the nuclei of the basal cells of normal prostatic glands stain with p63, while the malignant glands in prostatic adenocarcinoma (which lacks these cells) do not.[24] P63 is also helpful in distinguishing poorly differentiated squamous cell carcinoma from small cell carcinoma or adenocarcinoma. P63 should be strongly stained in poorly differentiated squamous cell, but negative in small cell or adenocarcinoma.[25]
Cytoplasmic staining on immunohistochemistry is seen in cells with muscle differentiation.[26]
Interactions
[edit]TP63 has been shown to interact with HNRPAB.[27] It also activates IRF6 transcription through the IRF6 enhancer element.[18]
Regulation
[edit]There is some evidence that the expression of p63 is regulated by the microRNA miR-203[28][29] and USP28 at protein level [30][31]
See also
[edit]- AMACR - another marker for prostate adenocarcinoma
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000073282 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000022510 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, et al. (September 1998). "p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities". Molecular Cell. 2 (3): 305–16. doi:10.1016/S1097-2765(00)80275-0. PMID 9774969.
- ^ Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, et al. (July 1998). "Cloning and functional analysis of human p51, which structurally and functionally resembles p53". Nature Medicine. 4 (7): 839–43. doi:10.1038/nm0798-839. PMID 9662378. S2CID 21953916.
- ^ Zeng X, Zhu Y, Lu H (February 2001). "NBP is the p53 homolog p63". Carcinogenesis. 22 (2): 215–9. doi:10.1093/carcin/22.2.215. PMID 11181441.
- ^ Tan M, Bian J, Guan K, Sun Y (February 2001). "p53CP is p51/p63, the third member of the p53 gene family: partial purification and characterization". Carcinogenesis. 22 (2): 295–300. doi:10.1093/carcin/22.2.295. PMID 11181451.
- ^ Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, et al. (May 2003). "DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development". Cancer Research. 63 (10): 2351–7. PMID 12750249.
- ^ Skipper M (January 2007). "Dedicated protection for the female germline". Nature Reviews Molecular Cell Biology. 8 (1): 4–5. doi:10.1038/nrm2091. S2CID 10702219.
- ^ Crum CP, McKeon FD (2010). "p63 in epithelial survival, germ cell surveillance, and neoplasia". Annual Review of Pathology. 5: 349–71. doi:10.1146/annurev-pathol-121808-102117. PMID 20078223.
- ^ Deutsch GB, Zielonka EM, Coutandin D, Weber TA, Schäfer B, Hannewald J, et al. (February 2011). "DNA damage in oocytes induces a switch of the quality control factor TAp63α from dimer to tetramer". Cell. 144 (4): 566–76. doi:10.1016/j.cell.2011.01.013. PMC 3087504. PMID 21335238.
- ^ Rouleau M, Medawar A, Hamon L, Shivtiel S, Wolchinsky Z, Zhou H, et al. (November 2011). "TAp63 is important for cardiac differentiation of embryonic stem cells and heart development". Stem Cells. 29 (11): 1672–83. doi:10.1002/stem.723. hdl:2066/97162. PMID 21898690. S2CID 40628077. Archived from the original on 2014-08-08.
- ^ Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. (July 2009). "TAp63 prevents premature aging by promoting adult stem cell maintenance". Cell Stem Cell. 5 (1): 64–75. doi:10.1016/j.stem.2009.04.003. PMC 3418222. PMID 19570515.
- ^ Roberts O, Paraoan L (Dec 2020). "PERP-ing into diverse mechanisms of cancer pathogenesis: Regulation and role of the p53/p63 effector PERP". Biochim Biophys Acta Rev Cancer. 1874 (1): 188393. doi:10.1016/j.bbcan.2020.188393. PMID 32679166. S2CID 220631324.
- ^ a b c Gebel J, Tuppi M, Sänger N, Schumacher B, Dötsch V (December 2020). "DNA Damaged Induced Cell Death in Oocytes". Molecules. 25 (23). doi:10.3390/molecules25235714. PMC 7730327. PMID 33287328.
- ^ Šimčíková D, Heneberg P (December 2019). "Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases". Scientific Reports. 9 (1): 18577. Bibcode:2019NatSR...918577S. doi:10.1038/s41598-019-54976-4. PMC 6901466. PMID 31819097.
- ^ a b c d e Dixon MJ, Marazita ML, Beaty TH, Murray JC (March 2011). "Cleft lip and palate: understanding genetic and environmental influences". Nature Reviews. Genetics. 12 (3): 167–78. doi:10.1038/nrg2933. PMC 3086810. PMID 21331089.
- ^ Shalom-Feuerstein R, Serror L, Aberdam E, Müller FJ, van Bokhoven H, Wiman KG, et al. (February 2013). "Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET". Proceedings of the National Academy of Sciences of the United States of America. 110 (6): 2152–6. Bibcode:2013PNAS..110.2152S. doi:10.1073/pnas.1201753109. PMC 3568301. PMID 23355677.
- ^ Qu J, Tanis SE, Smits JP, Kouwenhoven EN, Oti M, van den Bogaard EH, et al. (December 2018). "Mutant p63 affects epidermal cell identity through rewiring the enhancer landscape". Cell Reports. 25 (12): 3490–503. doi:10.1016/j.celrep.2018.11.039. hdl:2066/200262. PMID 30566872.
- ^ Qu J, Yi G, Zhou H (June 2019). "p63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes". Epigenetics & Chromatin. 12 (1): 31. doi:10.1186/s13072-019-0280-y. PMC 6547520. PMID 31164150.
- ^ a b Rotondo JC, Borghi A, Selvatici R, Magri E, Bianchini E, Montinari E, et al. (August 2016). "Hypermethylation-Induced Inactivation of the IRF6 Gene as a Possible Early Event in Progression of Vulvar Squamous Cell Carcinoma Associated With Lichen Sclerosus". JAMA Dermatology. 152 (8): 928–33. doi:10.1001/jamadermatol.2016.1336. PMID 27223861.
- ^ Shiran MS, Tan GC, Sabariah AR, Rampal L, Phang KS (March 2007). "p63 as a complementary basal cell specific marker to high molecular weight-cytokeratin in distinguishing prostatic carcinoma from benign prostatic lesions". The Medical Journal of Malaysia. 62 (1): 36–9. PMID 17682568.
- ^ Herawi M, Epstein JI (June 2007). "Immunohistochemical antibody cocktail staining (p63/HMWCK/AMACR) of ductal adenocarcinoma and Gleason pattern 4 cribriform and noncribriform acinar adenocarcinomas of the prostate". The American Journal of Surgical Pathology. 31 (6): 889–94. doi:10.1097/01.pas.0000213447.16526.7f. PMID 17527076. S2CID 9054387.
- ^ Zhang H, Liu J, Cagle PT, Allen TC, Laga AC, Zander DS (January 2005). "Distinction of pulmonary small cell carcinoma from poorly differentiated squamous cell carcinoma: an immunohistochemical approach". Modern Pathology. 18 (1): 111–8. doi:10.1038/modpathol.3800251. PMID 15309021.
- ^ Martin SE, Temm CJ, Goheen MP, Ulbright TM, Hattab EM (October 2011). "Cytoplasmic p63 immunohistochemistry is a useful marker for muscle differentiation: an immunohistochemical and immunoelectron microscopic study". Modern Pathology. 24 (10): 1320–1326. doi:10.1038/modpathol.2011.89. PMID 21623385.
- ^ Fomenkov A, Huang YP, Topaloglu O, Brechman A, Osada M, Fomenkova T, et al. (June 2003). "P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome". The Journal of Biological Chemistry. 278 (26): 23906–14. doi:10.1074/jbc.M300746200. PMID 12692135.
- ^ Yi R, Poy MN, Stoffel M, Fuchs E (March 2008). "A skin microRNA promotes differentiation by repressing 'stemness'". Nature. 452 (7184): 225–9. Bibcode:2008Natur.452..225Y. doi:10.1038/nature06642. PMC 4346711. PMID 18311128.
- ^ Aberdam D, Candi E, Knight RA, Melino G (December 2008). "miRNAs, 'stemness' and skin". Trends in Biochemical Sciences. 33 (12): 583–91. doi:10.1016/j.tibs.2008.09.002. hdl:2381/4113. PMID 18848452. Archived from the original on 2013-04-21.
- ^ Prieto-Garcia C, Hartmann O, Reissland M, Braun F, Fischer T, Walz S, et al. (Jun 2019). "The USP28-∆Np63 axis is a vulnerability of squamous tumours". bioRxiv: 683508. doi:10.1101/683508. S2CID 198263967.
- ^ Prieto-Garcia C, Hartmann O, Reissland M, Braun F, Fischer T, Walz S, et al. (March 2020). "Maintaining protein stability of ∆Np63 via USP28 is required by squamous cancer cells". EMBO Molecular Medicine. 12 (4): e11101. doi:10.15252/emmm.201911101. PMC 7136964. PMID 32128997.
Further reading
[edit]- Little NA, Jochemsen AG (January 2002). "p63". The International Journal of Biochemistry & Cell Biology. 34 (1): 6–9. doi:10.1016/S1357-2725(01)00086-3. PMID 11733180.
- van Bokhoven H, McKeon F (March 2002). "Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans". Trends in Molecular Medicine. 8 (3): 133–9. doi:10.1016/S1471-4914(01)02260-2. PMID 11879774.
- van Bokhoven H, Brunner HG (July 2002). "Splitting p63". American Journal of Human Genetics. 71 (1): 1–13. doi:10.1086/341450. PMC 384966. PMID 12037717.
- Brunner HG, Hamel BC, van Bokhoven H (October 2002). "P63 gene mutations and human developmental syndromes". American Journal of Medical Genetics. 112 (3): 284–90. doi:10.1002/ajmg.10778. PMID 12357472.
- Jacobs WB, Walsh GS, Miller FD (October 2004). "Neuronal survival and p73/p63/p53: a family affair". The Neuroscientist. 10 (5): 443–55. doi:10.1177/1073858404263456. PMID 15359011. S2CID 39702742.
- Zusman I (2005). "The soluble p51 protein in cancer diagnosis, prevention and therapy". In Vivo. 19 (3): 591–8. PMID 15875781.
- Morasso MI, Radoja N (September 2005). "Dlx genes, p63, and ectodermal dysplasias". Birth Defects Research. Part C, Embryo Today. 75 (3): 163–71. doi:10.1002/bdrc.20047. PMC 1317295. PMID 16187309.
- Barbieri CE, Pietenpol JA (April 2006). "p63 and epithelial biology". Experimental Cell Research. 312 (6): 695–706. doi:10.1016/j.yexcr.2005.11.028. PMID 16406339.
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, et al. (May 2011). "ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis". Cell Death and Differentiation. 18 (5): 887–96. doi:10.1038/cdd.2010.159. PMC 3131930. PMID 21127502.
External links
[edit]- TP73L+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- GeneReviews/NCBI/NIH/UW entry on Ankyloblepharon-Ectodermal Defects-Cleft Lip/Palate Syndrome or AEC Syndrome, Hay-Wells Syndrome. Includes: Rapp–Hodgkin Syndrome
- OMIM entries on AEC
- TP63 human gene location in the UCSC Genome Browser.
- TP63 human gene details in the UCSC Genome Browser.