Protein found in humans
| SMAD2 |
|---|
 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1DEV, 1KHX, 1U7V, 2LB3 |
|
|
| Identifiers |
|---|
| Aliases | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD family member 2, LDS6, CHTD8 |
|---|
| External IDs | OMIM: 601366 MGI: 108051 HomoloGene: 21197 GeneCards: SMAD2 |
|---|
| Gene location (Human) |
|---|
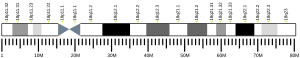
 | | Chr. | Chromosome 18 (human)[1] |
|---|
| | Band | 18q21.1 | Start | 47,808,957 bp[1] |
|---|
| End | 47,931,146 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 18 (mouse)[2] |
|---|
| | Band | 18 E3|18 51.42 cM | Start | 76,374,651 bp[2] |
|---|
| End | 76,444,034 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - Achilles tendon
- sperm
- secondary oocyte
- germinal epithelium
- monocyte
- islet of Langerhans
- amniotic fluid
- hair follicle
- endothelial cell
- sural nerve
|
| | Top expressed in | - saccule
- otic placode
- secondary oocyte
- endocardial cushion
- trigeminal ganglion
- ganglionic eminence
- nerve fiber layer
- sciatic nerve
- atrioventricular valve
- hand
|
| | More reference expression data |
|
|---|
| BioGPS | 
 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - phosphatase binding
- I-SMAD binding
- DNA-binding transcription factor activity
- DNA-binding transcription activator activity, RNA polymerase II-specific
- R-SMAD binding
- co-SMAD binding
- transcription factor binding
- metal ion binding
- RNA polymerase II cis-regulatory region sequence-specific DNA binding
- transforming growth factor beta receptor binding
- type I transforming growth factor beta receptor binding
- protein homodimerization activity
- chromatin binding
- protein binding
- double-stranded DNA binding
- SMAD binding
- DNA binding
- protein heterodimerization activity
- ubiquitin protein ligase binding
- primary miRNA binding
- disordered domain specific binding
- DNA-binding transcription factor activity, RNA polymerase II-specific
- tau protein binding
| | Cellular component | - cytoplasm
- cytosol
- nucleus
- heteromeric SMAD protein complex
- SMAD protein complex
- transcription regulator complex
- intracellular anatomical structure
- nucleoplasm
- activin responsive factor complex
- protein-containing complex
| | Biological process | - pattern specification process
- ureteric bud development
- endoderm development
- response to cholesterol
- organ growth
- embryonic pattern specification
- zygotic specification of dorsal/ventral axis
- post-embryonic development
- protein phosphorylation
- pericardium development
- regulation of binding
- transforming growth factor beta receptor signaling pathway
- negative regulation of cell population proliferation
- cell fate commitment
- common-partner SMAD protein phosphorylation
- regulation of transcription, DNA-templated
- SMAD protein signal transduction
- lung development
- signal transduction involved in regulation of gene expression
- insulin secretion
- in utero embryonic development
- negative regulation of transforming growth factor beta receptor signaling pathway
- negative regulation of gene expression
- nodal signaling pathway
- positive regulation of transcription, DNA-templated
- heart development
- pancreas development
- endoderm formation
- activin receptor signaling pathway
- SMAD protein complex assembly
- positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry
- embryonic foregut morphogenesis
- negative regulation of transcription by RNA polymerase II
- response to glucose
- positive regulation of epithelial to mesenchymal transition
- developmental growth
- gastrulation
- primary miRNA processing
- positive regulation of BMP signaling pathway
- embryonic cranial skeleton morphogenesis
- negative regulation of transcription, DNA-templated
- paraxial mesoderm morphogenesis
- intracellular signal transduction
- somatic stem cell population maintenance
- mesoderm formation
- regulation of transforming growth factor beta receptor signaling pathway
- positive regulation of gene expression
- anterior/posterior pattern specification
- positive regulation of transcription by RNA polymerase II
- transcription, DNA-templated
- transcription by RNA polymerase II
- protein deubiquitination
- wound healing
- adrenal gland development
- secondary palate development
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | |
|---|
NM_001003652
NM_001135937
NM_005901 |
| |
|---|
NM_001252481
NM_010754
NM_001311070 |
|
|---|
| RefSeq (protein) | |
|---|
NP_001003652
NP_001129409
NP_005892 |
| |
|---|
NP_001239410
NP_001297999
NP_034884 |
|
|---|
| Location (UCSC) | Chr 18: 47.81 – 47.93 Mb | Chr 18: 76.37 – 76.44 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
Mothers against decapentaplegic homolog 2, also known as SMAD family member 2 or SMAD2, is a protein that in humans is encoded by the SMAD2 gene.[5][6] MAD homolog 2 belongs to the SMAD, a family of proteins similar to the gene products of the Drosophila gene 'mothers against decapentaplegic' (Mad) and the C. elegans gene Sma. SMAD proteins are signal transducers and transcriptional modulators that mediate multiple signaling pathways.
Function
SMAD2 mediates the signal of the transforming growth factor (TGF)-beta, and thus regulates multiple cellular processes, such as cell proliferation, apoptosis, and differentiation. This protein is recruited to the TGF-beta receptors through its interaction with the SMAD anchor for receptor activation (SARA) protein. In response to TGF-beta signal, this protein is phosphorylated by the TGF-beta receptors. The phosphorylation induces the dissociation of this protein with SARA and the association with the family member SMAD4. The association with SMAD4 is important for the translocation of this protein into the cell nucleus, where it binds to target promoters and forms a transcription repressor complex with other cofactors. This protein can also be phosphorylated by activin type 1 receptor kinase, and mediates the signal from the activin. Alternatively spliced transcript variants encoding the same protein have been observed.[7]
Like other Smads, Smad2 plays a role in the transmission of extracellular signals from ligands of the Transforming Growth Factor beta (TGFβ) superfamily of growth factors into the cell nucleus. Binding of a subgroup of TGFβ superfamily ligands to extracellular receptors triggers phosphorylation of Smad2 at a Serine-Serine-Methionine-Serine (SSMS) motif at its extreme C-terminus. Phosphorylated Smad2 is then able to form a complex with Smad4. These complexes accumulate in the cell nucleus, where they are directly participating in the regulation of gene expression.
Nomenclature
The SMAD proteins are homologs of both the drosophila protein, mothers against decapentaplegic (MAD) and the C. elegans protein SMA. The name is a combination of the two. During Drosophila research, it was found that a mutation in the gene MAD in the mother repressed the gene decapentaplegic in the embryo. The phrase "Mothers against" was added, since mothers often form organizations opposing various issues, e.g., Mothers Against Drunk Driving, or (MADD). The nomenclature for this protein is based on a tradition of such unusual naming within the gene research community.[8]
Interactions
Mothers against decapentaplegic homolog 2 has been shown to interact with:
- ANAPC10,[9]
- DAB2,[10]
- EP300,[11][12]
- FOXH1,[11][13][14][15][16]
- HDAC1,[11]
- TGIF1,[11][12]
- Insulin receptor,[17]
- LEF1,[18]
- Myc,[19]
- MEF2A,[20]
- PIAS3,[21]
- PIN1,[22]
- SKI protein,[23][24]
- SKIL,[25][26]
- SMAD3,[27][28]
- SMURF2,[22][29][30]
- SNW1,[31]
- STRAP[32]
- WWTR1
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000175387 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000024563 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L (August 1996). "MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma". Cell. 86 (4): 543–52. doi:10.1016/S0092-8674(00)80128-2. PMID 8752209. S2CID 531842.
- ^ Riggins GJ, Thiagalingam S, Rozenblum E, Weinstein CL, Kern SE, Hamilton SR, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B (July 1996). "Mad-related genes in the human". Nat. Genet. 13 (3): 347–9. doi:10.1038/ng0796-347. PMID 8673135. S2CID 10124489.
- ^ "Entrez Gene: SMAD2 SMAD family member 2".
- ^ "Sonic Hedgehog, DICER, and the Problem With Naming Genes", Sep 26, 2014, Michael White. psmag.com
- ^ Nourry C, Maksumova L, Pang M, Liu X, Wang T (May 2004). "Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1". BMC Cell Biol. 5: 20. doi:10.1186/1471-2121-5-20. PMC 420458. PMID 15144564.
- ^ Hocevar BA, Smine A, Xu XX, Howe PH (June 2001). "The adaptor molecule Disabled-2 links the transforming growth factor β receptors to the Smad pathway". EMBO J. 20 (11): 2789–801. doi:10.1093/emboj/20.11.2789. ISSN 0261-4189. PMC 125498. PMID 11387212.
- ^ a b c d Wotton D, Lo RS, Lee S, Massagué J (April 1999). "A Smad transcriptional corepressor". Cell. 97 (1): 29–39. doi:10.1016/S0092-8674(00)80712-6. ISSN 0092-8674. PMID 10199400. S2CID 6907878.
- ^ a b Pessah M, Prunier C, Marais J, Ferrand N, Mazars A, Lallemand F, Gauthier JM, Atfi A (May 2001). "c-Jun interacts with the corepressor TG-interacting factor (TGIF) to suppress Smad2 transcriptional activity". Proc. Natl. Acad. Sci. U.S.A. 98 (11): 6198–203. Bibcode:2001PNAS...98.6198P. doi:10.1073/pnas.101579798. ISSN 0027-8424. PMC 33445. PMID 11371641.
- ^ Liu B, Dou CL, Prabhu L, Lai E (January 1999). "FAST-2 Is a Mammalian Winged-Helix Protein Which Mediates Transforming Growth Factor β Signals". Mol. Cell. Biol. 19 (1): 424–30. doi:10.1128/MCB.19.1.424. ISSN 0270-7306. PMC 83900. PMID 9858566.
- ^ Liu F, Pouponnot C, Massagué J (December 1997). "Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes". Genes Dev. 11 (23): 3157–67. doi:10.1101/gad.11.23.3157. ISSN 0890-9369. PMC 316747. PMID 9389648.
- ^ Dou C, Lee J, Liu B, Liu F, Massague J, Xuan S, Lai E (September 2000). "BF-1 Interferes with Transforming Growth Factor β Signaling by Associating with Smad Partners". Mol. Cell. Biol. 20 (17): 6201–11. doi:10.1128/MCB.20.17.6201-6211.2000. ISSN 0270-7306. PMC 86095. PMID 10938097.
- ^ Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M (September 1997). "Smad4 and FAST-1 in the assembly of activin-responsive factor". Nature. 389 (6646): 85–9. Bibcode:1997Natur.389...85C. doi:10.1038/38008. ISSN 0028-0836. PMID 9288972. S2CID 11927346.
- ^ O'Neill TJ, Zhu Y, Gustafson TA (April 1997). "Interaction of MAD2 with the carboxyl terminus of the insulin receptor but not with the IGFIR. Evidence for release from the insulin receptor after activation". J. Biol. Chem. 272 (15): 10035–40. doi:10.1074/jbc.272.15.10035. ISSN 0021-9258. PMID 9092546.
- ^ Labbé E, Letamendia A, Attisano L (July 2000). "Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and Wnt pathways". Proc. Natl. Acad. Sci. U.S.A. 97 (15): 8358–63. Bibcode:2000PNAS...97.8358L. doi:10.1073/pnas.150152697. ISSN 0027-8424. PMC 26952. PMID 10890911.
- ^ Feng XH, Liang YY, Liang M, Zhai W, Lin X (January 2002). "Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B)". Mol. Cell. 9 (1): 133–43. doi:10.1016/S1097-2765(01)00430-0. ISSN 1097-2765. PMID 11804592.
- ^ Quinn ZA, Yang CC, Wrana JL, McDermott JC (February 2001). "Smad proteins function as co-modulators for MEF2 transcriptional regulatory proteins". Nucleic Acids Res. 29 (3): 732–42. doi:10.1093/nar/29.3.732. PMC 30396. PMID 11160896.
- ^ Long J, Wang G, Matsuura I, He D, Liu F (January 2004). "Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3)". Proc. Natl. Acad. Sci. U.S.A. 101 (1): 99–104. Bibcode:2004PNAS..101...99L. doi:10.1073/pnas.0307598100. ISSN 0027-8424. PMC 314145. PMID 14691252.
- ^ a b Nakano A, Koinuma D, Miyazawa K, Uchida T, Saitoh M, Kawabata M, Hanai J, Akiyama H, Abe M, Miyazono K, Matsumoto T, Imamura T (March 2009). "Pin1 down-regulates transforming growth factor-beta (TGF-beta) signaling by inducing degradation of Smad proteins". J. Biol. Chem. 284 (10): 6109–15. doi:10.1074/jbc.M804659200. ISSN 0021-9258. PMID 19122240.
- ^ Harada J, Kokura K, Kanei-Ishii C, Nomura T, Khan MM, Kim Y, Ishii S (October 2003). "Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation". J. Biol. Chem. 278 (40): 38998–9005. doi:10.1074/jbc.M307112200. ISSN 0021-9258. PMID 12874272.
- ^ Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q (September 1999). "The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling". Genes Dev. 13 (17): 2196–206. doi:10.1101/gad.13.17.2196. ISSN 0890-9369. PMC 316985. PMID 10485843.
- ^ Stroschein SL, Bonni S, Wrana JL, Luo K (November 2001). "Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN". Genes Dev. 15 (21): 2822–36. doi:10.1101/gad.912901. ISSN 0890-9369. PMC 312804. PMID 11691834.
- ^ Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K (October 1999). "Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein". Science. 286 (5440): 771–4. doi:10.1126/science.286.5440.771. ISSN 0036-8075. PMID 10531062.
- ^ Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P (September 1997). "TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4". EMBO J. 16 (17): 5353–62. doi:10.1093/emboj/16.17.5353. ISSN 0261-4189. PMC 1170167. PMID 9311995.
- ^ Lebrun JJ, Takabe K, Chen Y, Vale W (January 1999). "Roles of pathway-specific and inhibitory Smads in activin receptor signaling". Mol. Endocrinol. 13 (1): 15–23. doi:10.1210/mend.13.1.0218. ISSN 0888-8809. PMID 9892009. S2CID 26825706.
- ^ Lin X, Liang M, Feng XH (November 2000). "Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling". J. Biol. Chem. 275 (47): 36818–22. doi:10.1074/jbc.C000580200. ISSN 0021-9258. PMID 11016919.
- ^ Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL (June 2001). "TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation". Nat. Cell Biol. 3 (6): 587–95. doi:10.1038/35078562. ISSN 1465-7392. PMID 11389444. S2CID 23270947.
- ^ Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP (May 2001). "Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-beta-dependent transcription". J. Biol. Chem. 276 (21): 18243–8. doi:10.1074/jbc.M010815200. ISSN 0021-9258. PMID 11278756.
- ^ Datta PK, Moses HL (May 2000). "STRAP and Smad7 Synergize in the Inhibition of Transforming Growth Factor β Signaling". Mol. Cell. Biol. 20 (9): 3157–67. doi:10.1128/MCB.20.9.3157-3167.2000. ISSN 0270-7306. PMC 85610. PMID 10757800.
Further reading
- Wrana JL (1998). "TGF-beta receptors and signalling mechanisms". Mineral and Electrolyte Metabolism. 24 (2–3): 120–30. doi:10.1159/000057359. PMID 9525694. S2CID 84458561.
- Massagué J (1998). "TGF-beta signal transduction". Annu. Rev. Biochem. 67: 753–91. doi:10.1146/annurev.biochem.67.1.753. PMID 9759503.
- Verschueren K, Huylebroeck D (2000). "Remarkable versatility of Smad proteins in the nucleus of transforming growth factor-beta activated cells". Cytokine Growth Factor Rev. 10 (3–4): 187–99. doi:10.1016/S1359-6101(99)00012-X. PMID 10647776.
- Wrana JL, Attisano L (2000). "The Smad pathway". Cytokine Growth Factor Rev. 11 (1–2): 5–13. doi:10.1016/S1359-6101(99)00024-6. PMID 10708948.
- Miyazono K (2000). "TGF-beta signaling by Smad proteins". Cytokine Growth Factor Rev. 11 (1–2): 15–22. doi:10.1016/S1359-6101(99)00025-8. PMID 10708949.
- Zannis VI, Kan HY, Kritis A, Zanni E, Kardassis D (March 2001). "Transcriptional regulation of the human apolipoprotein genes". Front. Biosci. 6: D456–504. doi:10.2741/Zannis. PMID 11229886.
PDB gallery
-
1dev: CRYSTAL STRUCTURE OF SMAD2 MH2 DOMAIN BOUND TO THE SMAD-BINDING DOMAIN OF SARA -
1khx: Crystal structure of a phosphorylated Smad2 -
1mjs: MH2 domain of transcriptional factor SMAD3 -
1mk2: SMAD3 SBD complex -
1u7f: Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex -
1u7v: Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex |
|
|---|
(1) Basic domains |
|---|
| (1.1) Basic leucine zipper (bZIP) | |
|---|
| (1.2) Basic helix-loop-helix (bHLH) | | Group A | |
|---|
| Group B | |
|---|
Group C
bHLH-PAS | |
|---|
| Group D | |
|---|
| Group E | |
|---|
Group F
bHLH-COE | |
|---|
|
|---|
| (1.3) bHLH-ZIP | |
|---|
| (1.4) NF-1 | |
|---|
| (1.5) RF-X | |
|---|
| (1.6) Basic helix-span-helix (bHSH) | |
|---|
|
|
(2) Zinc finger DNA-binding domains |
|---|
| (2.1) Nuclear receptor (Cys4) | | subfamily 1 | |
|---|
| subfamily 2 | |
|---|
| subfamily 3 | |
|---|
| subfamily 4 | |
|---|
| subfamily 5 | |
|---|
| subfamily 6 | |
|---|
| subfamily 0 | |
|---|
|
|---|
| (2.2) Other Cys4 | |
|---|
| (2.3) Cys2His2 | |
|---|
| (2.4) Cys6 | |
|---|
| (2.5) Alternating composition | |
|---|
| (2.6) WRKY | |
|---|
|
|
|
(4) β-Scaffold factors with minor groove contacts |
|---|
|
|
(0) Other transcription factors |
|---|
|
|
see also transcription factor/coregulator deficiencies |
|
|---|
| Type I | | ALK1 (ACVRL1) | - Kinase inhibitors: K-02288
- ML-347 (LDN-193719, VU0469381)
- Other inhibitors: Disitertide
|
|---|
| ALK2 (ACVR1A) | - Kinase inhibitors: DMH-1
- DMH-2
- Dorsomorphin (BML-275)
- K-02288
- ML-347 (LDN-193719, VU0469381)
|
|---|
| ALK3 (BMPR1A) | - Kinase inhibitors: DMH-2
- Dorsomorphin (BML-275)
- K-02288
|
|---|
| ALK4 (ACVR1B) | - Kinase inhibitors: A 83-01
- SB-431542
- SB-505124
|
|---|
| ALK5 (TGFβR1) | |
|---|
| ALK6 (BMPR1B) | - Agonists: BMP (2, 4, 5, 6, 7, 8A, 8B, 15 (GDF9B))
- Dibotermin alfa
- Eptotermin alfa
- GDF (5 (BMP14), 6 (BMP13), 7 (BMP12), 9, 15)
- Radotermin
- Kinase inhibitors: DMH-2
- Dorsomorphin (BML-275)
- K-02288
|
|---|
| ALK7 (ACVR1C) | - Antagonists: Lefty (1, 2)
- Kinase inhibitors: A 83-01
- SB-431542
- SB-505124
|
|---|
|
|---|
| Type II | | TGFβR2 | - Kinase inhibitors: DMH-2
- LY-364947
|
|---|
| BMPR2 | |
|---|
| ACVR2A (ACVR2) | - Agonists: Activin (A, B, AB)
- BMP (2, 4, 5, 6, 7, 8A, 8B, 15 (GDF9B))
- Dibotermin alfa
- Eptotermin alfa
- GDF (1, 3, 5 (BMP14), 6 (BMP13), 7 (BMP12), 9, 11 (BMP11), 15)
- Myostatin (GDF8)
- Nodal
- Radotermin
|
|---|
| ACVR2B | - Agonists: Activin (A, B, AB)
- BMP (2, 4, 6, 7)
- Dibotermin alfa
- Eptotermin alfa
- GDF (1, 3, 5 (BMP14), 6 (BMP13), 7 (BMP12))
- Myostatin (GDF8)
- Nodal
- Osteogenin (BMP3, BMP3A)
- Radotermin
- Decoy receptors: Ramatercept
|
|---|
| AMHR2 (AMHR) | |
|---|
|
|---|
| Type III | |
|---|
| Unsorted | |
|---|
This article incorporates text from the United States National Library of Medicine, which is in the public domain.

 1dev: CRYSTAL STRUCTURE OF SMAD2 MH2 DOMAIN BOUND TO THE SMAD-BINDING DOMAIN OF SARA
1dev: CRYSTAL STRUCTURE OF SMAD2 MH2 DOMAIN BOUND TO THE SMAD-BINDING DOMAIN OF SARA 1khx: Crystal structure of a phosphorylated Smad2
1khx: Crystal structure of a phosphorylated Smad2 1mjs: MH2 domain of transcriptional factor SMAD3
1mjs: MH2 domain of transcriptional factor SMAD3 1mk2: SMAD3 SBD complex
1mk2: SMAD3 SBD complex 1u7f: Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex
1u7f: Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex 1u7v: Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex
1u7v: Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex